yasmeenzeena.github.io
Translation
Main principles and key players of protein translation
mRNA —> amino acids in polypeptide chain </br>
transcriptome in humans = 100,000 mRNAS </br> Proteome in humans = 400,000 proteins </br>
this discrepancy is due to alternative splicing, where one mRNA precursor molecule can make multiple different proteins from one single gene by splicing them differently and arranging the exons in new ways. [polycistronic mRNAs are known to exist in eukaryotic viruses: Ryabova LA, Pooggin MM, Hohn T. Viral strategies of translation initiation: ribosomal shunt and reinitiation, Prog Nucleic Acid Res Mol Biol , 2002, vol. 72 (pg. 1-39)& in drosophila but not yet found in humans.]
Gene expression is governed by extracellular signals which result in a protein being transcribed and translated.

mRNAs: carrier of information in translation; remember uses U not T!; 64 possible codons, 3 of which are stop codons (UAA, UGA, UAG); Methionine is the common start codon AUG -> defines the beginning of the Kozac sequence in eukaryotes. Shine-Dalgarno, the prokaryotic sequence, is a ribosome binding site that occurs 8 bases upstream of the AUG codon.
All amino acids apart from methionine and tryptophan have multiple codons coding for them = degenerate genetic code.
Kozac sequence: (gcc)gccRccAUGG
Shine-Dalgarno (SD) Sequence is a ribosomal binding site in bacterial and archaeal messenger RNA, generally located around 8 bases upstream of the start codon AUG: AGGAGG.
Map of the mRNA pre translation:
5’- 7-methyl guanosine cap —> 5’-UTR —> coding region from start AUG codon to stop codon –> 3’ UTR–> polyA tail -‘3
The guanosine is not coded for by the DNA but added on after transcription, it is methylated on the 7 position directly after capping in vivo by a methyltransferase. Capping protects from degradation, allows functionality.
The polyA tail allows the mRNA to circularise, which is needed in translation. It allows Rho independent termination to halt transcription in prokaryotes and helps maintain RNA stability.
tRNAS: 75-95 bases long, about 100 types in eukaryotes –> all have a CCA 3’ terminal sequence on the acceptor stem where amino acids get added. The loop opposite the 3’ and 5’ ends is the anticodon loop, which gives the tRNA its specificity to bind to a specific codon.
the tRNA+amino acid structure = aminoacyl-tRNA
aminoacyl tRNA synthetase = + enzyme that matches the amino acid with its correct tRNA molecule + 20, one for each amino acid + forms an aminoacyl-tRNA + amino acid can now be added to the peptide via aminoacyl tRNA transferase + enzyme features an editing site to ensure that the right tRNA has been added
aminoacyl tRNA transferases = enzymes that take the amino acid from the aminoacyl-tRNA and adds it to the polypeptide chain
These enzymes must be able to distinguish between tRNAs -> via the acceptor arm, which has stem and 3’end regions specific to each tRNA. The discriminator base proceeding the 3’ CCA end (position 73) is particularly useful in identification of the right tRNA!
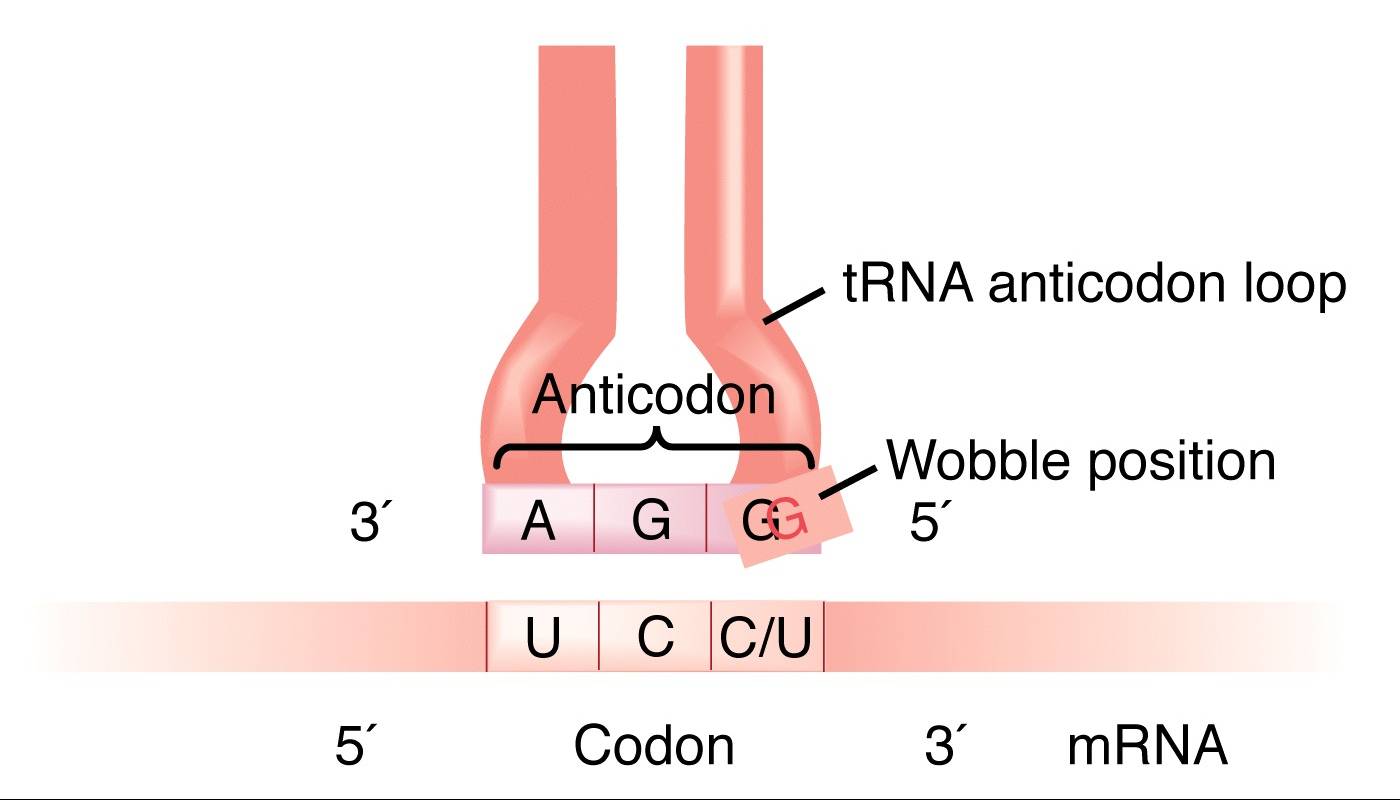
As there are less than 61 tRNAs there is not one tRNA for each codon, so non-Watson Crick base pairing often occurs between the codon and the anticodon. Results in wobble between the 3’ base of the mRNA and the 5’ base of the tRNA anticodon.
Ribosomes: protein synthesising machines in the cytoplasm
+ able to elongate polypeptide chains at a rate of 3-5 amino acids per second
+ ribosomes are structural molecules lacking enzymes but have "ribozymes" in the large subunit with catalytic functions
+ ribosomes have both RNA and protein in their structure
+ ribosomes move along the mRNA during translation
+ 80S complex is made up of subunits
+ ribosomes are only made when there is a demand for protein synthesis, otherwise they ar degraded
+ ribosomes are made in the nucleolus and moved out into the cytoplasm
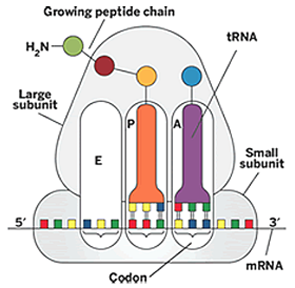
+ ribosomes hold the mRNA and the tRNAs together during translation:
mRNA binds to the small subunit, the P site in the ribosome interacts with the tRNA carrying the growing polypeptide chain, the A site interacts with the tRNA carrying the next amino acid to be added.

Translation initiated differently in prokaryotes and eukaryotes!
Prokaryotes = Shine Dalgarno/Ribosome Binding Site -> found 5-9 bases upstream of the start AUG codon, there is where the 16S subunit of the ribosome will bind to the mRNA.
Eukaryotes = Kozac sequence -> 5’CCACCAUGG 3’ found around the start methionine. This is not the ribosome binding site however, that occurs either in the 5’ cap or as an internal ribosome entry site (IRES.)
[In a few bacterial mRNAs, GUG is used as the initiator codon, and CUG occasionally is used as an initiator codon for methionine in eukaryotes & some mitochondria read UGA, the stop codon, as a Trp codon p133 lodish.]
Polysomes: complex of an mRNA strand where multiple ribosomes are able to translate the strand at the same time. mRNA is circularised here to allow this process. This allows many copies of the protein to be made from one mRNA strand.
The fate of newly translated proteins:
+ translocation, via a leader sequence informing the cell of the protein destination (e.g. transmembrane proteins shuttled to the plasma membrane via exocytosis pathways, see protein trafficking lectures.)
+ post translational modification in ER and Golgi -> this includes disulphide bridges in the ER!!!!
-
activation e.g. phosphorylation in Golgi: creates a dock in the molecule -> works in signalling pathways! + Free OH needed (serine, threonine, tyrosine), turned into the PO4 by protein kinase + e.g. lipid kinase can phosphorylate things on the membrane (where it sits)
-
degradation (via ubiquitination) + mono ubiquitination or can add multiple molecules / chains of ubiquitin + ubiquitin - 76aa protein, very stable + added to lysine residues on proteins by ubiquitin ligase + e.g. STAT1 is phosphorylated, translocates to the nucleus and is ubiquitinated -> undergoes proteosome mediated degradation ???????
-
Coacetylation: covalent binding of coenzyme A -> metabolism target, modification for protein in stress response + CoA can covalently bind to proteins when the cell is under stress (e.g. starvation) + during normal conditions there is very little protein CoAcetylation + in stress conditions CoA is added to proteins -> added to the cystine residues by covalent linkage + protect the cysteine residues from oxidation to suphenic acid (very reactive under stress) + thus can think of CoA as an anti oxidant modification in the cell!
-
acetylation regulates histone proteins: closed chromatin —> relaxed chromatin if you add an acetyl group, increases transcription + histone tails are required for chromatin to condense from the beads-on-a-string conformation into the 30 nm fiber + histone tails are positive due to K and R residues + Histone tails undergo reversible acetylation and deacetylation by enzymes that act on specific lysines in the N-termini + acetyl groups cancel out the positivity of the lysines + chromatin forms the “beads on a string” conformation due to this neutralisation + DNA is in the open conformation -> ready for transcription
-
methylation of histone proteins: + Condensed regions of chromatin known as heterochromatin are often methylated heavily + heterochromatin cannot easily be unwound for transcription -> allows silencing of genes
Molecular mechanism of translation
In prokaryotes transcription and translation are continuous with each other due to a lack of compartmentalisation within the cell. Their mRNA is usually polycistronic and will contain multiple ribosome start and stop codons, whilst eukaryotic mRNA is spliced into one monocistronic transcript.
Initiation: rate limiting, once initiation has begun it cannot be stopped so it is very tightly controlled. ATP and GTP hydrolysis is needed, and after initiation a complex containing the ribosome, mRNA and initiator Met-tRNA is formed. Eukaryotic translation initiation factors (eiFs) help mediate the complex formation. Many bind GTP, and the GTP-GDP hydrolysis is treated as a proofreading measure, such that complexes before GTP hydrolysis are unstable and can reform, thus allowing fixing of any errors in the structure.
2 possible mechanisms used:
- Cap-dependent initiation: 5’ cap structure is scanned until the AUG is found
- Internal ribosome entry site (IRES): initiation complex binds upstream of the AUG codon, used when the cap isn’t available e.g. cap production halted if certain cellular stressors applied.
Normal eukaryotic translation uses cap dependent initiation:
- Circularisation:
- eIF4A and B have helicase activity and unwind any RNA secondary structure
- eIF4E [eukaryotic initiation factor 4E] binds to the the 5’ cap
- eIF 4G binds to the polyA-binding protein, PABP, which is already bound to the polyA tail (helps the formation of the polyA tail in mRNA processing) + the eIF 4G acts as a scaffold onto which other factors can bind
- eIF4E and eIF4G are associated, so the mRNA is forced to circularise
- mechanism ensures that the mRNA has a cap and a tail!

- 43S complex is made and translation is initiated
- tRNA with the methionine bound is bound to eIF2
- 40S ribosomal subunit is bound by eIF1, eIF1A, eIF5, eIF3 and eIF2-tRNA-methionine + eIF1A and eIF3 promote binding of the eIF2-tRNA-methionine (ternary) complex to the 40S subunit
- the mRNA-eIF4 complex then associates with the 43S complex through an interaction between eIF4G and elF3: now the mRNA, the start amino acid and the ribosome are brought together

- once the Kozac sequence and the AUG are found another binding event occurs:
- [Lodish: eIF2 GTP is hydrolysed when AUG is encountered, irreversible step which presents further scanning. As the methionine is base paired onto the mRNA the complex is now called a 48S complex.]
- the 60S subunit is added on the other side of the mRNA to sandwich it in and hold it in place as the ribosome scans the rest of the sequence.
- 60S is recruited by the eIF5, which acts as the final proofreader. 60S is added by GTP hydrolysis: once hydrolysis occurs the final 80S assembly has been made
- All of the eukaryotic initiation factors are removed before elongation begins
Elongation
The key steps in elongation are entry of each succeeding aminoacyl-tRNA with a tRNA, formation of a peptide bond, and the translocation of the ribosome one codon at a time along the mRNA.
Ribosome A site: binding site for the tRNA with the amino acid </br> Ribosome P site: binding site for the tRNA lacking an amino acid i.e. deacylated. the P-site holds the tRNA which is linked to the growing polypeptide chain </br> Ribosome E site: exit site, tRNA leaves from here </br>
At the beginning, methionine-tRNA is bound to the P site on the assembled 80S ribosome, so the next aminoacyl-tRNA binds to the A site. EF1A-GDP is turned to EF1A-GTP by EF1B. EF1A-GTP brings the aminoacyl-tRNA to the A site via a GTP hydrolysis. A peptide bond is formed between the methionine and the next amino acid. The eEF2 hydrolyses it’s GTP an the conformational change results in it moving the the deacetylated tRNA (previously with methionine) from the P site to the E site, and moving the tRNA with the bound peptide to the P site from the A site. A new aminoacyl-tRNA will bind to the A site via EF1A, make a peptide bond to the second amino acid, and be moved to the P site by eEF2.
- first tRNA is in the P site with the bound peptide, new aminoacyl-tRNA binds to the A site (EF1A)
- peptide bond catalysed: peptide now on the tRNA in the A position
- translocation occurs to get the peptide back to the P position:
- old deacylated tRNA moves P->E and tRNA now bound to the peptide moves A->P by eEF2

aminoacyl-tRNA binding —> peptide bond catalysis —> translocation
Alberts: Translating an mRNA molecule. Each amino acid added to the growing end of a polypeptide chain is selected by complementary basepairing between the anticodon on its attached tRNA molecule and the next codon on the mRNA chain. Because only one of the many types of tRNA molecules in a cell can base-pair with each codon, the codon determines the specific amino acid to be added to the growing polypeptide chain. The four-step cycle shown is repeated over and over during the synthesis of a protein. In step 1, an aminoacyl-tRNA molecule binds to a vacant A site on the ribosome. In step 2, a new peptide bond is formed. In step 3, the large subunit translocates relative to the small subunit, leaving the two tRNAs in hybrid sites: P on the large subunit and A on the small, for one; E on the large subunit and P on the small, for the other. In step 4, the small subunit translocates carrying its mRNA a distance of three nucleotides through the ribosome. This “resets” the ribosome with a fully empty A site, ready for the next aminoacyl-tRNA molecule to bind. As indicated, the mRNA is translated in the 5ʹ-to-3ʹ direction, and the N-terminal end of a protein is made first, with each cycle adding one amino acid to the C-terminus of the polypeptide chain.
Termination
The stop codons do not have a specific tRNA! Instead, releasing factors aka eRFs are used:
eRF1 = similar shape to tRNAs, acts by binding to the ribosomal A site and recognising stop codons directly eRF3 = GTP-binding protein. Promotes cleavage of the peptidyltRNA, thus releasing the completed protein chain
The peptidyl-tRNA bond of the tRNA in the P site is not cleaved, terminating translation, until one of the three stop codons is correctly recognised by eRFl, another example of a proofreading step in protein synthesis.
- eRF1 binds to the stop codon in the ribosomal A site (mimics tRNA)
- it’s catalytic activity is stimulated by eRF3
- nucleophilic attack on the ester bond between the peptide and the P site tRNA
- cleavage of the peptidyltRNA
- releases the completed protein chain
eRF1 is very similar in shape to tRNA, conversed: anticodon loop (domain 1 in eRF1) </br> aminoacyl acceptor stem (domain 2) </br> T stem of a tRNA molecule (domain 3) </br>
Proofreading
tRNAs have many copies so we need a high affinity of interaction to get the right tRNA to the right codon sequence. Mismatches tRNAs dissociate from the A site before GTP hydrolysis of eEF1A can occur -> the aminoacyl-tRNA from the ternary complex does not remain bound to the ribosomal A site but dissociates.
GTP hydrolysis, and hence tight binding, does not occur if the anticodon of the incoming aminoacyl-tRNA cannot basepair with the codon at the A site. In this case, the ternary complex diffuses away, leaving an empty A site that can associate with other aminoacyl-tRNA-Eflex·GTP complexes until a correctly base-paired tRNA is bound. Thus, GTP hydrolysis by EF1A is another proofreading step that allows protein synthesis to proceed only when the correct amino acylated tRNA is bound to the A site. This phenomenon contributes to the fidelity of protein synthesis.
When is the IRES pathway used? When the 5’ caps are either damaged or unavailable e.g. in starvation the cell does not produce global induction of proteins but instead must selectively translate only survival proteins. IRES is used to select only these proteins -> quick & specialised regulatory response. Often hijacked by viruses either to stop translation (picornoviruses) or to make viral proteins (hepatitis C.)
picornoviruses is able to inactivate IRES -> IRES is depend on the C-terminal fragment of eIF4G to recruit the 40S subunit through interaction with eIF3 -> picornoviruses can cleave eIF4 to inactive translation
Hepatitis C -> needs only eIF3 to start IRES, once hepatitis C binds it’s mRNA’s AUG is placed into the P site via a large conformational change of the 40S ribosome subunit.
Other viral targets:
Puromycin - Aminoacyl-tRNA analog, causes premature chain termination </br> Tetracycline - Inhibits aminoacyl-tRNA binding to the A site </br> Erythromycin - binds peptidyl transferase and blocks polypeptide translocation </br>