yasmeenzeena.github.io
L11&12: Protein Trafficking
Subcellular compartmentalisation and the problem of getting proteins to the right place i.e. protein trafficking. Proteins encoded by nuclear genes begin synthesis in the cytosol. Proteins destined to remain in the cytosol have no sorting signals (the default pathway): targeting to other destinations requires sorting signals.
“protein trafficking = signal-based targeting + vesicle based trafficking
signal based targeting: cytoplasm -> specific organelle e.g. membrane proteins get inserted into the plasma bilayer, or for hydrophilic proteins allows insertion into an organelle through the bilayer. destination = ER, mitochondria, golgi, etc!
vesicle-based trafficking secretory pathway: proteins from the ER to destination via vesicles; all proteins start in the ER so any protein in the secretory pathway must be initially targeted to the ER -> nascent proteins, aka still being synthesised on the ER, are extruded directly onto the ER membrane -> translocate across -> assemble in their native form (catalysts are present in the ER lumen, 1/3 of all native folding in the cell happens here) -> post translational modifications occur -> proteins moved out to another destination.
Proteins whose final destination is the Golgi, lysosome, plasma membrane, or cell exterior are transported along the secretory pathway by the action of small vesicles that bud from the membrane of one organelle and then fuse with the membrane of another
ER, mitochondria, nucleus, chloroplast, peroxiome = signal based targeting(??);
Quick overview of sorting: Fig. 13-1 p578 Lodish 7th edition 1. All nuclear-encoded mRNAs are translated on cytosolic ribosomes 2. non secretory pathways: proteins that remain in the cytosol don’t have an ER signal. Their synthesis occurs on free ribosomes 3. Proteins with a secretory signal are released into the cytosol too, but then are moved to their respective organelle 4. the nucleus, mitochondria and chloroplast are still considered non secretory pathways as the protein doesn’t leave the cell. 5. Secretory pathway: Ribosomes synthesising nascent proteins in the secretory pathway are directed to the rough endoplasmic reticulum (ER) by an ER signal sequence 6. After translation is complete & post translational modifications & native folding, they go to the Golgi body 7. more post translational modification, then further sorting to the lysosome or the plasma membrane via vesicle transport.
Each organelle carries a set of receptor proteins that bind only to specific kinds of signal sequences: protein is translocated across the membrane via a translocation channel (unidirectional, protein cannot slide back into cytoplasm, coupled to ATP.GTP)
SIGNAL SEQUENCES:

The information to target a protein to a particular organelle destination is encoded within the amino acid sequence of the protein itself, usually within sequences of about 20 amino acids, known generically as signal sequences aka uptake-targeting sequences or signal peptides. usually at the N terminus (can be C or interior tho)
Usually at N (or C) terminus - one example of a “signal patch” where the amino acids only come together in the 3D folded protein
Signal sequences often are removed from the mature protein by specific proteases once translocation across the membrane is completed. Don’t have to be removed however!
How does a protein cross or become inserted into a physical barrier (ie. a lipid bilayer)?
Protein with it’s signal sequence needs to cross the bilipid layer: macromolecule, can’t diffuse. Needs:
- receptor on the membrane to recognise a signal
- channel to guide the protein through
- both channel and receptor made of protein
Nuclear protein import and export. See Lodish, pg. 615-621, Alberts, pg. 649-657
Nucleus = double membrane, have nuclear pores that span the inner and outer membranes & extend into the nucleus.
Nuclear pore complex: made of many copies of about 30 different nucleoporin proteins -> octagonal, membrane-embedded ring structure that surrounds a largely aqueous pore. Three types of nucleoporins:
- structural nucleoporins: scaffold of the nuclear pore, seven of these make a Y complex aka the basic unit of the scaffold. Some of the scaffold nucleoporins are structurally related to vesicle coat protein complexes: suggests a common evolutionary origin for NPCs and vesicle coats.
- membrane nucleoporins:
- FG nucleoporins: line the channel, contain multiple repeats rich in phenylalanine and glycine. FG repeats are thought to have two main functions: interacting weakly with each other to create a gel like tangle which halts macromolecular diffusion of proteins above 40kDa through the pore, and also as dock sites for nuclear transport receptors.
Nuclear pores are considered gated channels for larger polypeptides that require active transport through the channel.
Histones, transcription factors, and DNA and RNA polymerases = cytoplasm syntehsis and import to nucleus through nuclear pore complexes.
Nuclear-localisation signal (NLS) anywhere in the protein sequence direct transport into the nucleus: usually contains positive amino acids e.g. arginine and lysine. Occurs anywhere in the sequence so forms “loops” or “patches” on the protein surface. Many function even when linked as short peptides to lysine side chains on the surface of a cytosolic protein, suggesting that the precise location of the signal within the amino acid sequence of a nuclear protein is not important.
Import:
Nuclear pores are large so the polypeptide can be completely folded upon entry. This is why the nuclear localisation signal can be found in the 3D structure as a “patch.” NLS in the amino acid sequence are recognised by a soluble receptor in the cytoplasm = importin from the karyopherin family. It can bind to the phenylalanine-glycine repeats, which disrupts their weak interactions with each other, resulting in the gel like protein tangle of the pore locally disintegrating, allowing the protein through the channel.
Importin binds to the NLS in the cytoplasm –> takes the protein to the nuclear pore complex –> binds to the phenylalanine-glycine repeats –> protein tangle of the FG interactions disintegrates locally –> protein can pass through to the nucleus
Export:
NLS in export proteins are usually lysine rich but otherwise less defined. The soluble export receptor in the nucleus is the exportin, also of the karyopherin family.
Directionality:
Ran is a molecular switch, a small monomeric G protein that exists in either GTP- or GDP-bound conformations.
Ran in import: The importin-protein complex meets Ran-GTP once it enters the nucleus, which binds to displace the protein. The importin-Ran.GTP complex is removed to the cytosol where Ran-GAP hydrolyses the GTP into GDP. Ran.GDP returns to the nucleus via the NTF2 receptor. The GDP is converted to GTP via Ran-GEF (guanine exchange factor), ready for another round.
Ran in export: The Ran-GTP binds to exportin and the protein (triple complex.) This is transported out of the nucleus and into the cytosol, where Ran-GAP hydrolyses the GTP to GDP. The complex disassociates. Exportin is transferred back to the nucleus.
-
Ran-GAP is present in the cytosol, turns GTP -> GDP to hydrolyse protein complexes (either the Ran-GTP.importin or the Ran-GTP.exportin.protein)
-
Ran-GEF is present in the nucleus, and turns GDP -> GTP to allow new protein complexes to form (Ran-GTP.importin or the Ran-GTP.exportin.protein)
- Ran-GAP: breaks the complexes in the cytosol
- Ran-GEF: generates the complexes in the nucleus
Ran-GDP is high in the cytosol due to Ran-GAP presence, whereas Ran-GTP (needed to form protein complexes) has a high concentration in the nucleus due to Ran-GEF.
Ran is a molecular switch that moves things out of the nucleus into the cytoplasm: either moves importin back into the cytoplasm to attach to another protein, or moves the exported protein out of the nucleus.

Summary:
Import 1. Importin binds to the cargo protein in the cytoplasm. 2. The cargo complex is able to interact with the FG nucleoporins in the nuclear pore and diffuse through. 3. In the nucleoplasm, Ran·GTP binds to the importin and causes the cargo protein to unbind. 4. Importin is returned to the cytoplasm by the Ran.GTP in a complex which is hydrolysed by Ran-GAP in the cytoplasm to free the importin receptor. 5. Ran-GDP returns to the nucleus and is made into Ran-GTP by Ran-GEF
Export 1. Protein for removed is is bound to the exportin and Ran.GTP. 2. The exportin.protein.Ran·GTP complex is transported back to the cytoplasm. 3. Ran-GAP hydrolyses GTP -> GDP 4. Conformation change results in dissociation of the exportin. 5. Exportin and Ran·GDP return to the nucleus. 6. GEF causes formation of Ran-GTP.
In the nucleus = Ran-GTP due to GEF action In the cytoplasm = Ran-GDP due to GAP action
Protein import into mitochondria
Mitochondrial proteins are first fully synthesised as mitochondrial precursor proteins (unfolded) in the cytosol and then translocated into mitochondria by a post-translational mechanism.
Protein cannot be folded when entering the channel! Protein only folds in its native 3D shape in the matrix. Has proteins bound to stop it from folding until it reaches the mitochondrial membrane. Proteins are kept unfolded by Hsp70, a cytosolic ATPase protein. This chaperon protein prevents aggregation and spontaneous folding. They are stripped off as the protein enters the TOM complex.
Matrix targeting sequence is: AMPHIPATHIC α HELIX: N terminus with positive amino acids on one side, uncharged on the other side of the helix e.g. cytochrome oxidase, alcohol dehydrogenase. All outer membrane proteins have internal signal sequences which are not cleaved!
TOM and TIM transporters are situated where the double membranes are close together to allow translation into the matrix. TOM = access across the outer membrane into the inner mitochondrial space TIM (22/23) = access across the inner membrane into the matrix
All proteins must go through TOM. Proteins that end up in the outer membrane then go through the SAM complex. TIM23 directs proteins to the matrix or helps to insert transmembrane proteins into the inner membrane. Tim22 = more inner membrane proteins. The OXA complex directs the 13 proteins which are encoded for by the mitochondria itself. [SAM = Sorting and Assembly Machinery; OXA = cytochrome OXidase Activity; TIM = Translocator of the Inner Mitochondrial membrane; TOM = Translocator of the Outer Membrane.]
Other destinations than the matrix:
-
outer mitochondrial membrane e.g. porin molecules: goes through TOM in the outer membrane, bound by chaperons in the inner membrane space, goes through the SAM complex in the outer membrane
-
inner mitochondrial membrane (A): goes through the TOM and partially through TIM -> once signal sequence has gone through TIM is it cleaved –> the stop transfer sequence is exposed in the IMS —> hydrophobic so wants to stay in the membrane -> protein moves laterally into the inner mitochondrial membrane
-
inner mitochondrial membrane (B): protein goes to matrix via TOM and TIM –> cleavage of the signal sequence reveals the adjacent hydrophobic signal sequence at the new N terminus –> travels back up through the OXA transporter to get back to the IMM [this is the route that all 13 mitochondrial proteins take]
-
intermembrane space protein (C): through TOM, into TIM, stuck in the IMM by the hydrophobic sequence, which is cleaved by a protease –> free protein floats around in the IMS i.e. not anchored to the membrane.

Folds in the matrix via Hsp60 chaperone!
Energy
Used to get the get the polypeptide into the TOM transporter and unbind the Hsp70 proteins. Used to get the polypeptide into the TIM transporter. Used to get the polypeptide into the matrix. 1. cytosolic Hsp70 is released at the TOM complex via ATP hydrolysis 2. the inner membrane space –> matrix translocation via the TIM transporter is aided by the electrochemical gradient of the inner mitochondrial membrane 3. mitochondrial Hsp70 helps the polypeptide chain enter the matrix via ATP hydrolysis: either by binding and pulling the polypeptide, or but halting backslide through the TIM transporter.
Protein import into the ER: this can be either co- or post-translational
ER membrane is continuous with the nucleus membrane! ER = synthesis of membrane lipids & assembly of membrane proteins. all soluble proteins that will be secreted are directed here first = “secretory pathway” rough ER: bit of the ER membrane that receives proteins of the secretory pathway, very heavily ribosome studded
Secretory cells (e.g. pancreatic acinar cells) contain the organelles of the secretory pathway (e.g., ER and Golgi) in great abundance: used in research. pulse labelling experiment, radiolabled amino acids added to proteins synthesised on membrane bound ribosomes -> shows that these proteins are translocated across the membrane.
rough ER breaks up into closed vesicles with ribosomes studded on the outside upon homegenisation, aka rough microsomes -> still able to carry out protein translation so can be easily studied! hard to isolate the entire ER structure (too delicate) -> Labeling experiments demonstrate that secretory proteins are localised to the ER lumen shortly after synthesis: radiolabeled amino acids make labeled polypeptides, cells are homogenised, labeled polypeptides found in the microsomes, (easily separated from homogenate due to ribosomes, different density), microsomes treated with protease and some with detergent -> protease can only breakdown labeled polypeptides if detergent is present to break down the microsome bilayer, shows that polypeptides must be in the microsome. microsome = lumen of the ER. ^^ shows that polypeptides are swiftly moved into the lumen of the ER from the ribosome.
16- to 30-residue ER signal sequence in the N terminus of the nascent protein directs the ribosome to the ER membrane and initiates translocation: all have one or more + charged amino acids adjacent to 16+ hydrophobic amino acids aka hydrophobic core -> the core if deleted the N terminus cannot function as a signal, protein remains in the cytosol (can be added to cytosolic proteins to make them translocate!) -> core is the binding site
Co-translational translocation
(most eukaryotes): the transport of most secretory proteins into the ER lumen begins while the incompletely synthesised (nascent) protein is still bound to the ribosome:
(ribosomes on the outside of the ER are there because they are co-translocating proteins into the ER lumen!)
cytosolic ribonucleoprotein particle, binds to ER signal sequence in nascent protein and the large ribosomal subunit = complex, which then binds to the SPR receptor on the ER membrane
1. ER signal sequence translated by the ribosome
2. ER signal sequence bound by SRP (via the P54 M domain and the hydrophobic core of the nascent protein's hydrophobic interactions)
3. Pause in translation
4. SRP delivers the ribosome/nascent polypeptide complex to the SRP receptor in the ER membrane
5. GTP binds to both P54 and the a subunit of the ER receptor
6. ribosome/nascent polypeptide transferred to Sec61, protein complex forming a channel in the ER membrane
7. As the polypeptide chain elongates, it passes through the translocon channel into the ER lumen
8. In the lumenthe signal sequence is cleaved by signal peptidase and is rapidly degraded
- SRP => 6 proteins bound to a 300bp RNA (hexamer scaffold.) P54, one of the protein domains, binds to the ER signal sequence on the nascent protein. The M domain of P54 has hydrophobic amino acids (e.g. meth) –> the hydrophobic core of the signal peptide binds to this cleft via hydrophobic interactions. Notable as RNA is associated! not purely protein.
- SRP receptor => integral protein of the ER membrane, two subunits: a and b. Binding with the SRP-nascent protein complex is best when P54 and a subunit both are GTP bound. In bacteria, P54 and the a subunit homologue come together to form a heterodimer, whose active site is able to bind and hydrolyse two GTPs (neither active site is well suited to bind a single GTP alone.)
GTP hydrolysis results in transfer of the ribosome/nascent polypeptide to Sec61, a protein complex that forms a channel in the ER membrane. (The ribosome is also moved!!! remember.) SRP dissociates and cycles back to the cytosol. As translation continues, the elongating chain passes directly from the large ribosomal subunit into the central pore of Sec61. The 60S subunit is aligned so that the chain never encounters the cytosol and thus only folds in the ER lumen.
As the growing polypeptide chain enters the lumen of the ER, the signal sequence is cleaved by signal peptidase, which is a transmembrane ER enzyme -> allows native folding of protein once translated in the ER lumen.

Co-translational translocation summary:
• signal = extreme N terminus, positively charged amino acid (arginine/lysine) with a hydrophobic sequence following. Cleaved in the ER by signal peptidase enzyme.
• receptor part one = SRP, protein:RNA complex in cytosol
• receptor part two = membrane SRP receptor on the ER membrane
• polypeptide nascent chain's N terminus is bound by the SRP, pause in translation -->
• taken to SRP receptor on the ER membrane, threaded through the Sec61 channel complex into the ER lumen
• polypeptide is synthesised into the ER
• The signal sequence is eventually cleaved by signal peptidase
Post-translational translocation: yeast! proteins enter only once translation is complete!
Proteins must be kept in their unfolded state by chaperone proteins! e.g. Hsp70 in the cytosol, using ATP.
Same Sec61 but no SRP/SRP receptor bringing the ribosome/protein complex to the Sex61. There is a direct interaction between the Sec61 and the signal sequence in the protein instead -> the unidirectional aspect of translocation is provided by Sec63 complex and BiP, a molecular chaperon, member of the Hsc70, tetrameric.
BiP = in the ER lumen, Sec63 = embedded in the membrane. BiP has a peptide-binding domain and an ATPase domain. BiP’s role is to stop the protein from backsliding through Sec61 into the cytosol.
- Once the N terminal segment of the protein enters the ER lumen, signal peptidase cleaves the signal sequence just as in cotranslational translocation 2. BiP.ATP + Sec63 interact = hydrolysis of bound ATP, conformational change in BiP results in BiP binding to the polypeptide chain. 3. As more of the polypeptide enters the ER lumen it successively binds to multiple BiPs, stopping backsliding back into the cytosol. Polypeptide continues into the ER lumen studded with ADP molecules from the BiP binding. 4. BiP molecules spontaneously exchange their bound ADP for ATP, leading to release of the polypeptide, which can then fold into its native conformation. (kinda similar to mitochondrial import!)
ER import summary:
- Via Sec61 complex translocon
- Requires signal sequences: one or more charged aa followed by hydrophobic region
- Can be co- or post-translational
- Proteins must be unfolded via Hsp70 chaperon proteins to pass through Sec61
- Co-translational uses SRP and SRP receptor on the ER membrane, GTP hydrolysis pushes the polypeptide through the Sec61 translocon
- Post-translation uses Sec63 and BiP to provide unidirectional transport (ATPase)
- Co-translational import requires GTP, post-translational import requires ATP
N-linked glycosylation in the ER
Addition of sugars via enzymes: oligosaccharide transferase, membrane protein of the ER membrane that binds to the ribosome, recognises any asparagine residues coming through -> asparagine something serine/thrionine. Adds a sugar onto the asp: some N-acetylglucosamine, then some mannose, then glucose.
Glycosylation seems to assist folding & stability and can play a role in cell cell adhesion at the cell surface. ^N linked / core glycosylation -> first type to occur! happens at asparagine.
Protein folding in the ER and the dislocation or retrotranslocation of misfolded proteins (ERAD)
Chaperones help proteins to fold correctly and also mark incorrectly folded proteins for degradation

The ER is also the site of disulphide bond formation(S-S). Disulphide bonds (between cysteine residues) help stabilise the tertiary and quarternary structureof proteins. Con only form in the oxiding environment of the ER (cytosol is reducing!)
ERAD: ER Associated Degradation -> protein is misfolded or marked as wrong, must be removed, sent out through a translocon (unknown which) into the cytosol, sugar is removed, ubiquitin is added (poly ubiquitination), marked for degradation! Process of removal is called retrotranslocation.
Insertion of membrane proteins into the ER membrane
Signal sequence moves through Sec61 and is cleaved by signal peptidase as per usual. A hydrophobic region is now stuck in Sec61 so moves laterally into the membrane, rest of the protein is attached! = membrane protein anchored in place.
Can also make a transmembrane protein like this e.g. rhodopsin -> hydrophobicity plot shows that the hydrophobic regions will transfer latterly into the membrane: each hydrophobic region will form a transmembrane region. Force of translation pushes the hydrophilic bits into the ER lumen. 7 membrane spanning domains.
Protein sorting and trafficking beyond the ER.
The ER is the starting place for proteins destined for many other locations. We will discuss these routes through the cell: secretion and the secretory pathway; endocytosis and the endocytic pathway; and retrograde or recycling pathways
ER resident proteins remain there e.g. BiP, chaperon proteins etc.
Secretory pathways: from ER to cell surface or lysosome Endocytic pathways: from cell surface into the cell! Recycling pathways: backwards transport pathway to balance, takes things away from the cell surface to keep it from growing too large.
Signals on the proteins: 3D patches, sequences, or even protein modifications can direct traffic e.g. glycosylation,ubiquitination, lipid modifications All processes from the ER involves something other than translocation! Must use vesicle based trafficking now.
Vesicular transport, and how proteins may be sorted into different vesicles. This section includes vesicle coats and the role they play in forming the vesicle and selecting the proteins to be included in it (“cargo” proteins). See Lodish 634-636 or Alberts pg. 697-699.
Vesicles can form from an organelle: have coats, e.g. clathrin, COPI and COPII. Protein coats around the vesicles.
COPI&II can sort which proteins go into the vesicles. Clathrin has a coat component and adaptors, accessory proteins that do the selection of proteins. each coat needs a GTPase for binding assembly.
Targeting signals direct proteins to the correct location
As seen previously, two methods to move proteins from the ER to another organelle: vesicular transport, or direct fusion to create a hybrid organelle (e.g. late endosome and lysosome fusion.) Vesicular transport works using coat proteins, which are able to select the right proteins to be added to the vesicle. -> The first step in vesicular transport is the formation of a vesicle by budding from the membrane.
Clathrin = coat protein, moves molecules from the plasma membrane i.e. endocytosis, and moves from the Golgi to the endosome. [ARF is a recruiting protein, Hsp70 needed to uncoat.]
COPI = bud from cis Golgi to ER, recycling pathway [ARF is the GTPase needed to uncoat]
COPII = bud from the ER to the Golgi, secretory pathway [Sar1 is the GTPase needed to uncoat.]
The assembly of vesicle coats also requires GTP-binding proteins, which appear to regulate the binding of coat proteins to the membrane. The budding of COPI-coated vesicles from the Golgi complex requires a GTP-binding protein called ARF (ADP-ribosylation factor), while the budding of COPII-coated vesicles from the ER requires a distinct GTP-binding protein called Sar1.
The clathrin coat is made up of clathrin and accessory proteins, which buds on the cytosolic facing membrane to allow the molecules to enter the cell/endosome. The process is described below:
- Coat assembly and cargo selection: The adaptor proteins bind to the cargo receptors on the membrane and recruit clathrin to the membrane. They decide which proteins will be added to the ligands.
- Bud formation: Adaptor proteins are able to attract the necessary proteins. These proteins come together and introduce curvature into the membrane due to their large number, resulting in a bud forming. Receptor and cargo taken up into the bud.
- Vesicle formation: neck of the bud is severed by other membrane bending and fission proteins, a vesicle is formed.
- Uncoating: the coat is rapidly lost once the vesicle forms (via ATPase of the Hsp70 family.) Clathrin and adaptor removed. Proteins of interest are in the transport vesicle -> binds to the endosome, first target of the endocytosis pathway.

Specific membrane fusion: SNAREs, and different types of membrane fusion
After formation of a vesicle is completed, the coat is shed, exposing the vesicle’s v-SNARE proteins (membrane proteins.) v-SNARES have associated tethering proteins such as Rab-GTP. Rab-GTP is able to bind to a Rab effector, a tethering protein, which brings the vesicle closer to the target membrane [tethering]. v-SNARES and t-SNARES on the target membrane can bind together. t-SNARES contribute 3 alpha helicases from either 2 or 3 t-SNARES to bind to the single v-SNARE from the vesicle. A four helical bundle is made now [docking.] This can also be called a a “trans SNARE pair”. It allows the two membranes to fuse together, mixing their lipids and transferring the soluble proteins in the vesicle to the organelle lumen.
Rab-GTP + Rab effector = tethering v-SNARE + t-SNARE = docking, vesicle and target membrane fusion
The SNARE complex (trans-SNAR-pair) must be removed before membrane fusion by an ATPase complex called NSF, a six subunit protein with an associated alpha-SNAP molecule. This complex is a universal way to remove SNAREs, as contrasted to the many different types of v and t SNARES possible. ATP is used to remove the SNARE complex.
homotypic fusion = one compartment with itself e.g. early endosome + early endosome
heterotypic fusion = different membranes fusing together
^ same mechanism for both
The sorting of both integral-membrane and soluble ER resident proteins
All proteins start synthesis on the ER, co translocated into the ER.
Soluble ER resident protein (e.g. BiP): retrieval pathway
COPII vesicles will take some of the fluid from the lumen of the ER each time it buds, thus some protein will escape and end up in the cis-Golgi. ER resident proteins contain the KDEL targeting signal on their C terminus, which is recognised by the KDEL receptor in the cis-Golgi membrane. cis-Golgi has a lower pH to the ER, allowing binding of the protein to the KDEL receptor. COPI takes the protein back to the ER (recycling pathway.) Once back in the ER the KDEL protein dissociates from the receptor due to the neutral pH environment.
COPII + protein —> cisGolgi protein + KDEL receptor bind (low pH) COPI + protein + KDEL receptor —-> ER protein + KDEL receptor unbind (neutral pH)

Transmembrane ER resident proteins
Membrane protein must stay in the ER. Taken by COPII budding by accident (part of the membrane is removed each time a vesicle is made), goes to the cis-Golgi. Membrane protein has a signal: two lysines & two of any amino acid at the extreme C terminus (KKXX.) KKXX interacts with the coat proteins of COPI, concentrated into the COPI vesicles, returned to the ER via a retrieval pathway.
COPII + protein –> cisGolgi KKXX signal interacts with COPI coat proteins COPI + protein –> ER
Secretion, including Golgi protein modifications (glycosylation, sulfation).
ER -> via COPII -> cisGolgi –> medial –> transGolgi –> (transGolgi network) –> constitutive secretion to the plasma membrane soluble proteins will be released out of the cell (exocytosis) whilst membrane proteins will be added to the plasma membrane
all cells do constitutive secretion, some specialised ones do regulated secretion (membrane + soluble proteins held at the plasma membrane in regulatory vesicles, only fuse once a signal has been sent e.g. transmitter release, hormone secretion (e.g. insulin waits until blood glucose level lowered), digestive enzymes.
Golgi modifications: O-linked glycosylation occurs in the Golgi (unlike the N linked glycosylation of the ER) -> happens on OH groups in serine/threonine where sugars are added to the hydroxyl groups.
Two models for proteins getting through the Golgi:
1. Vesicular transport method: from each compartment a vesicle is budded off and fuses with the next compartment along
2. Cisternal maturation model: favoured model, idea is that a compartment matures into the next one, so no vesicle action. cisGolgi becomes medial, medial matures intro trans etc. After the trans-Golgi complex vesicles bud off to go to the next organelle. Maturation works via "back transport" -> enzymes go backwards over time e.g. enzymes of the transGolgi travel back to the medial, which results in the medial becoming the transGolgi. Each compartment 'moves up one' going right to left.

The sorting of soluble lysosomal proteins: the mannose-6-phosphate system
Function is to degrade proteins and lipids, concentrated enzymes! fuses with the late endosome to degrade these proteins. most acidic organelle. organelles across the protein maturation pathway slowly get more acidic!
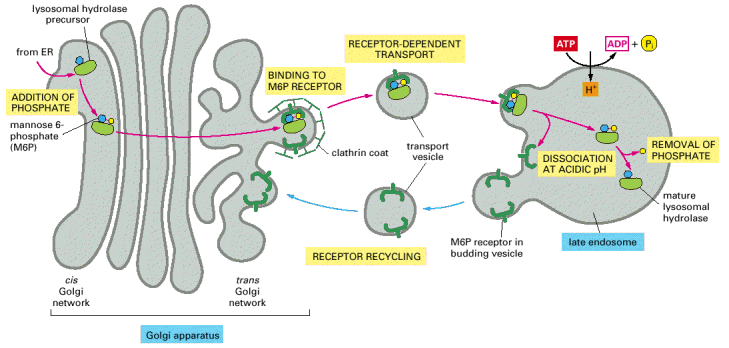
Hydrolase enzymes must be brought to the lysosome -> sugar signal, mannose 6 phosphate added to the protein in the Golgi (phosphorylated in the Golgi) -> signal bound by the mannose 6 phosphate receptor in the late Golgi —> coated vesicle transports protein to the late endosome -> acidic organelle due to H+ pumped in, dissociation of receptor and enzyme —> receptor is recycled by COPI back to the Golgi –> enzyme added into the lysosome once the endosome and lysosome fuse together in direct fusion
The LDL receptor
Membrane protein, synthesised on the ER, taken to the plasma membrane by constitutive secretion.
LDL receptor is on the plasma membrane and binds LDL, using the FDNPVY signal (endocytosis signal.) The adaptor proteins bind to the receptor and protein, and clathrin is recruited to form a vesicle. Budding, vesicle formation, Hsp70 uncoats –> vesicle fuses with the endosome, the acidic environment causes the LDL and the receptor to dissociate. The receptor is recycled back to the plasma membrane. The LDL remains in the endosome until direction fusion with the lysosome occurs. LDL is broken down to free cholesterol, which goes into the cytoplasm.
EGF receptor
EGF can cause cancer if left switched on too long = epidermal growth factor.
EGF binds to the EGF receptor at the plasma membrane using a endocytosis signal: tyrosine -X-X-hydrophobic amino acid. EGF is taken up and ubiquitin is EGF receptor during endocytosis –> goes to the endosome. The EGF-receptor-ubiquitin complex is NOT dissociated but transported to the late endosome. This compartment can bud inwards, so EGF complex is sorted into the inner membrane by escort proteins tat target the ubiquitin. EGF complex ends up in the intra-luminal vesicles (ubiquitin might be removed before this stage?) –> signal is switched off! cytosolic part of the receptor is no longer at the cytosol so the signal cannot be maintained –> when endosome fuses with lysosome everything is degraded.
The invagination processes are essential for complete digestion of endocytosed membrane proteins: because the outer membrane of the multivesicular body becomes continuous with the lysosomal membrane, which is resistant to lysosomal hydrolases; the hydrolases, for example, could not digest the cytosolic domains of endocytosed transmembrane proteins, such as the EGF receptor shown here, if the protein were not localised in intralumenal vesicles.