yasmeenzeena.github.io
Nucleic Acids
A = deoxyadenosine G = deoxyguanosine C = deoxycytidine T = deoxythymidine
deoxyadenosine-5’-triphosphate = dATP deoxyguanosine-5’-triphosphate = dGTP deoxycytidine-5’-triphosphate = dCTP deoxythymidine-5’-triphosphate = dTTP
Nucleases cleave phosphodiester bonds:
- Endonuclease - cleaves bonds internally (all restriction sites at once)
- Exonuclease - starts at one end (exo) of a polynucleotide chain and cleaves bonds one at a time
- can have different polarities, ie either cleaves from 5-3 or 3-5
Restriction endonucleases (type II)
cleaves both strands of duplex DNA at restriction sites, which are palindromic. Cuts can be staggered (sticky ends) or blunt. {if enzymes are not handled using their optimum conditions, you can get unexpected offtarget effects, called “star activity”. Star activity can include anything from cleavage at incorrect sites to single base substitutions, and can be caused by many factors, including extended incubation, too much glycerol, or not enough magnesium.}
enzyme EcoR1 (Escherichia coli R1) cuts 5’-G/AATTC-3’ and leave 5’ overhangs (i.e. the 5’ strand is the short one, the 3’ strand has the longer sticky end.)
enzyme Pst1 (Pseudomonas stuartii 1) cleaves along the 5′-CTGCA/G-3′ sequence, leaving 3’ overhangs.

enzyme Hpa1 (Haemophilus parainfluenzae 1) creates a blunt end by cleaving the 5’- GTT/AAC-3’ site.
DNA ligase
= joins up DNA by catalysing the formation of a phosphodiester linkage between the 5’- phosphate and the 3’-OH molecules of adjacent nucleotides in DNA.
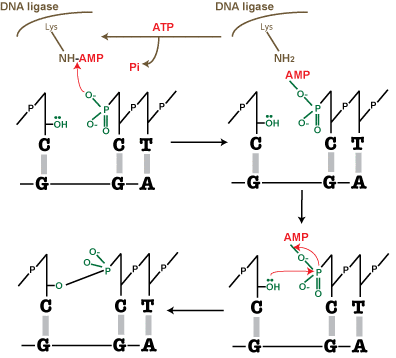
Mechanism of ligation by DNA ligase:
ATP hydrolysis (favourable) is coupled to the energetically unfavourable sealing of the DNA nick.
- ATP + DNA ligase = DNA-ligase-AMP and PP
- Ligase must be adenylated: AMP is added onto a lysine residue on the enzyme [functional group is ((CH2)4NH2).] The positively charged NH3+ on the lysine functional group binds to the AMP, which takes the place of one of the hydrogen bonds. Meanwhile the two phosphates are expelled.
- DNA-AMP molecule is formed. The NH(+)-AMP has a positive charge, which allows it to be attacked by the negative oxygen on the 5’ phosphate of the nicked nucleotide.
- AMP is transferred from DNA ligase to the nucleotide: AMP-phosphate bond.
- This AMP-phosphate bond is attacked by the 3′-OH of the nucleotide on the other side of the gap. The lone pair on the O of the OH attacks the phosphate which is now delta+.
- The phosphate-OH bond is covalent and the AMP-phosphate bond breaks.
- The nick is sealed
- AMP is released
- DNA ligase needs a new source of AMP to function again
note: ATP is the source of AMP in eukaryotes, viruses & phage ligases, but NAD+ is the source in bacterial ligase.
Alkaline phosphatase:
- Removes the 5’ phosphate group from DNA/RNA
- DNA without a 5’-phosphate cannot ligase (no phosphodiester linkage can form)
- Done to prevent self ligation of the DNA
- However the 3’-OH ends are still intact, which allows the vector to bind to the recombinant DNA via its 5’-phosphate
- The second strand will NOT be able to ligase however, leaving a small gap. This gap will be corrected by the enzyme repair mechanisms in the host bacterial cell once the recombinant vector is transformed.
DNA melting/hyperchromicity
When double-stranded ‘ordered’ DNA goes to a single-stranded ‘disordered’ structure the Absorbance (A) at 260 nm increases. This is because the bases become unstacked andcan absorb more light. Hydrogen bonds, which interfere with aromatic resonance, are also broken. Nuclear bases are better chromophores when they are “loose” (ie no hydrogen bonds or stacking) as in single stranded DNA because their aromatic resonance is less hindered.
Tm = melting point of DNA i.e. when 50% of the duplex DNA has become single stranded through denaturing (~80 Celsius.) Found at the halfway point of absorbance260 increase. Tm depends on the sequence composition -> A&T rich DNA denatures faster because of the two hydrogen bonds, whilst G&C have three.
Topoisomerase type I
Enzyme that relaxes supercoiled DNA. DNA becomes overwound ahead of the replication fork as the replication bubble moves along. Torsion to the DNA strand is abated by topoisomerases nicking the backbone of one of both strands.
Agarose Gel
Supercoiled DNA travels the fastest along the agarose gel -> followed by linear DNA –>
followed by open/circular DNA
Supercoiled = in vivo plasmid DNA, desired type of DNA.
Linear = DNA helix has been cut in both strands to open out the DNA. Migration
predictable by the molecular weight of the DNA.
Nicked/circular = DNA that has been nicked by a topoisomerase enzyme, usually because
of a nearby replication fork
Ethidium bromide as an intercalating agent: added to agarose gel to show where the DNA bands are. The chemical is able to slide in between the bases as it is planar aromatic but gets stuck there and can cause mutations. Its aromatic structure makes it a chromophore that fluoresces in UV light. GelRed is a non mutagenic version used in labs to avoid cancer risks