yasmeenzeena.github.io
L8: Bacterial gene control
Aims of this lecture:
- The bacterial operon:
- Positive and negative regulation of the lactose operon
- Regulation of the tryptophan operon by a repressor protein and by the process of attenuation.
- Two-component regulatory systems – phosphate concentration and gene regulation.
- Quorum sensing and the lux operon – you will PROBABLY not find this information in most textbooks. The notes given should suffice.
- The ribosomal and transfer RNA operons of E. coli
Positive and negative regulation of the lactose operon
Housekeeping proteins are always present in the cell in a constant concentration, but other proteins are linked to nutrient amounts in the environment.
Genes coding for proteins involved in same metabolic pathway are clustered into an OPERON! Therefore a cluster of genes can make one long mRNA which is later split up into the individual proteins its codes for “polycistronic mRNA.”

The polycistronic mRNA will have a number of AUGs (methoinine = start codon) and UGAs (stop codon) to indicate where each polypeptide begins and ends in the RNA. When the mRNA is being translated the ribosome will bind to AUG, translate til UGA and unbind. This will occur for each polypeptide in the RNA sequence.
Genes are only transcribed when needed!
inducible operons = must be turned on
repressible operons = must be turned off
Lac Operon
Inducible operon = usually occur in a catabolic pathway (i.e. breakdown) e.g. sugar breakdown, lac operon. Activated by a sugar molecule!
- glucose (breaks the β- galactoside linkage)
- Lactose: inducer, sugar that binds preferentially to the promoter so that the LacI
- repressor cannot bind. Its actually allolactose that binds, an isomer of lactose (made by the low levels of β- galactosidase that are always in the cell!)
- Glucose: sugar that is metabolised preferentially to lactose via catabolite repression: glucose is phosphorylised over adenylyl cyclase, enzyme inactive so cAMP cannot be made to activate CAP, no CAp = down transcription of genes as RNA poly binding is weak
- RNA polymerase: transcribes the Z, Y & A genes, cannot bind if the LacI repressor protein is bound to the promoter/operator region first. Needs the help of the catabolite activator protein (CAP) to bind.
- LacI: makes the repressor protein, as its own promoter so is continuously expressed.
- LacZ: β- galactosidase
- LacY: lactose permease (moves lactose into the cell)
- LacA: thiogalactoside transacetylase (helps remove toxic metabolites e.g. thiogalactosides, which are transported out of cell by lac Y protein)
If the operon is repressed, the LacI gene produces its repressor protein which binds to the operator (which overlaps with the promoter) and stops the RNA polymerase enzyme transcribing the LacZ, Y and A genes. The operator region is a negative regulatory site as it stops transcription. The RNA polymerase has a weak affinity to the DNA because the lac promoter is weak, and so needs the help of the catabolite activator protein (CAP) to bind.
The lac operator sequence of the O1 (primary operator) is a near perfect inverted repeat, i.e. there are two half sites on either side with the repeat. One molecule of repressor protein can bind to each half site = 2 repressor molecules bound at O1. There is also an auxiliary operator upstream of the O1 before the promotor which can also bind two repressor proteins. The four repressor proteins are arranged in a tetramer, which forces the DNA strands to curve around the tetramer to allow binding to O1 and O3. There is another operator sequence much further downstream that has also been shown to form a loop with the primary O1.

If lactose is present the operon is activated! An isomer of lactose, allolactose, binds to the repressor protein and causes a conformational shift which results in the repressor dissociating from the operator region.
The lac operon is usually turned OFF i.e. negative control as the bacteria prefer to use glucose over lactose: if glucose is present the lac operon transcription is inhibited [glucose metabolism doesn’t need new proteins whilst lactose metabolism requires Z and Y and A so transcription and translation needed: energy conserved.]
Glucose inhibits the lac operon, so that if lactose and glucose are both present there is no transcription of the genes! This is achieved through catabolite repression i.e. a glucose metabolite inhibits transcription (which proves the glucose metabolism is occurring and the cell doesn’t need lactose.)
When glucose enters the cell the first stage of glycolysis is it converted into glucose-6- phosphate. Since the glucose is phosphorylated the enzyme adenylyl cyclase is not, so it remains inactive. This enzyme can also make cAMP by adding a P to ATP, but this reaction only occurs when glucose is not present. Thus, increase in [glucose] = decrease in [cAMP].

These cAMP molecules are able to bind to the catabolite activator protein and activate it, which allows it to bind to DNA. When cAMP is low, less CAP is activated and the RNA polymerase cannot bind. Thus, high glucose is able to inhibit the lac operon being transcribed.
The CAP protein increase the affinity of RNA polymerase for the weak lac operon. The CAP binding to the CAP site on the DNA causes the DNA to bend 90 degrees, which increases the affinity for RNA polymerase. The CTD (carboxyl terminal domain) of the RNA polymerase binds to the CAP protein now that the DNA is bent.
Lactose paradox: how can lactose induce the operon if the lacY gene is needed to transport lactose into the cell initially? A: The lac repressor is a highly specific DNA binding protein that binds very tightly to O1 and O2. But no protein can bind forever. When it dissociates, an mRNA is made and some permease and some β-galactosidase is synthesized. The repressor quickly rebinds and transcription is blocked.
The effect of this “escape” synthesis is that there will always be a few molecules of permease and a few molecules of β-galactosidase inside the cell. When the cell encounters lactose in the medium enough can be taken up and converted to allolactose to induce the operon. http://sandwalk.blogspot.co.uk/2008/10/lactose-paradox.html
SUMMARY:
-
Lactose present, no glucose: cAMP binds to CAP, making CAP able to bind DNA Bound CAP helps RNA polymerase attach to the lac operon promoter Lactose in cell via lactose permease Lactose –> allolactose via β-galactosidase Allolcatose binds to repressor preferentially RNA polymerase can now transcribe!
-
Only glucose: High glucose means low cAMP as adenylyl cyclase is involved in the first stage of glycolysis not cAMP production Lack of cAMP = no activation of catabolite activator protein, CAP RNA polymerase cannot bind to the weak promoter No transcription of Z Y A genes
-
No sugars: lac operon is not activated cAMP is high so CAP is activated but repressor from LacI is bound to the operator site
Regulation of the tryptophan operon by a repressor protein and by the process of attenuation (repressible operators)
Transcription is turned off when enough of the product is made in a cell. e.e. trp operon, involved in tryptophan. Transcription of trp is high when [trp] is low, but drops off once enough is made. The trp operon is expressed (turned “on”) when tryptophan levels are low and repressed (turned “off”) when they are high. https://www.khanacademy.org/science/biology/gene-regulation/gene-regulation-inbacteria/ a/the-trp-operon

The operon here involves five genes (E to A are structural) with a single promoter and operator. Between the operator and the first gene is the leader sequence. The leader encodes a short polypeptide and also contains an attenuator sequence. Overall regulation happens in two ways: through the attenuator sequence (which is transcribed into mRNA) or through a repressor binding.
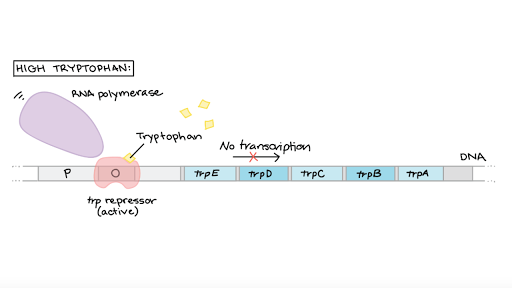
When try is present in high enough quantities the trp repressor binds to the operon and stops RNA polymerase from transcribing the genes, thus switching off the cell’s ability to make more trp. Here trp is considered a corepressor because it binds to the trp repressor and activates it, allowing it to bind to the operon.
The trp repressor is encoded by a trp R gene, which is not located in the trp operon & has it’s own promoter. The protein it makes is called a “apo-repressor” aka needs a corepressor to function.
In low trp concentration, the trp repressor is not activated and will not bind. In this system, the trp repressor acts as both a sensor and a switch. It senses whether tryptophan is already present at high levels, and if so, it switches the operon to the “off” position.
Attenutation is another mechanism to stop trp production - it stops the completion of the mRNA and halts transcription. Once the leader strand is transcribed into mRNA it can be translated immediately (prokaryote host!) and it makes 2 try residues and the attenuator sequence.
If trp conc is high the ribosome translates all of leader mRNA region one quickly and stops at the stop codon in region two. As region two is covered by the ribosome regions 3&4 hydrogen bond together, creating a hairpin loop structure. This is a Ro independant terminator which results in the dissociation of RNA polymerase. The structural genes cannot be transcribed now!

When trp is low the ribosome binds at methionine but is stopped at the trp codons and waits for tRNA. This allows region 2 to hydrogen bond to region 3 –> stops 3&4 bonding (4 hasn’t been transcribed yet) so no hairpin loop structure forms. The RNA polymerase is not dissociated and the ribosome continues along.
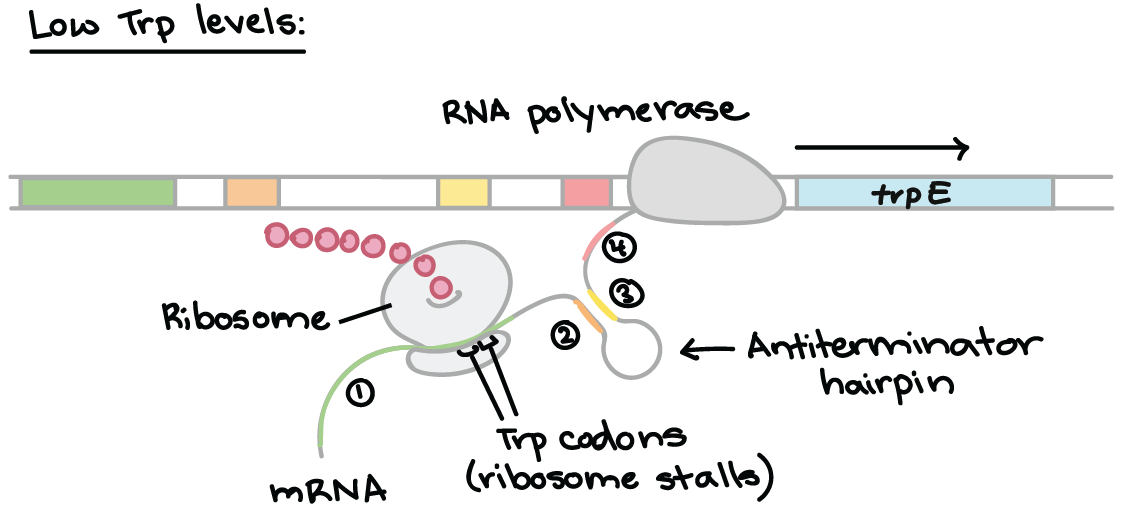
If the leader mRNA sequence is translated quickly, because the tRNA can bring the trp resides over immediately due to their high intracellular concentration, the ribosome will not pause at all. . aka Rho independent termination.
- RNA poly has just made mRNA out of the leader and attenuator strand, is at the structural genes
- Ribosome attaches right after and beings to translate the mRNA
- It reaches the trp codons in the leader mRNA first
- Low trp levels:
- tRNA takes a while to bring the trp, ribosome pauses in the middle of region one
- this allows regions 2&3 to hydrogen bond together
- no hairpin loop structure is made
- once the trp residues are added ribosome continues normally
- High trp levels:
- the tRNA brings the trp quickly
- ribosome only pauses at the stop codon in region 2 of the leader mRNA
- region two is blocked so region 3 hydrogen bonds to region 4, causing a hairpin loop structure to form
- release of RNA polymerase
- structural genes cannot be transcribed
Two component regulatory systems - phosphate concentration and gene regulation
Phosphate operon: has a sensor protein (PhoR) and a regulatory protein (PhoB.)
PhoR has three domains: periplasmic (binds to the phosphate), transmembrane & cytoplasmic. When the [phosphate] decreases, the periplasmic domain releases its phosphate -> conformational change, the histidine on the cytoplasmid domain is exposed. The histidine is phosphorylated by ATP and the protein is now in its active sensor form.
Once PhoR has been activated it transfers its phosphate onto the aspartate of PhoB, the regulator protein. Active PhoB can now bind to DNA, increasing the affinity of the RNA polymerase for various promoters: genes phoA (alkaline phosphatase) phoS (phosphate binding protein) phoE (porin) & gpB (phosphate transporter.) PhoB binds near the promoter of each gene involved (~30 overall.) PhoB = transcription factor!
Quorum sensing and the lux operon
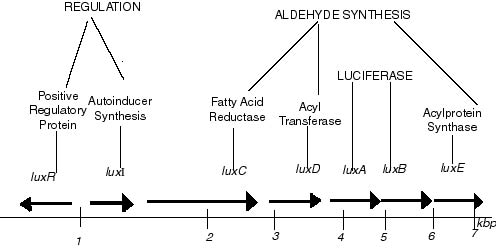
Vibrio fischeri are a bacterium that are able to bioluminesce when in a quorum (i.e. large numbers.) They often colonise in squid organs and other sea creatures. Each bacterium makes one molecule of VAI (Vibrio fischeri autoinducer) so a quorum will contain many of these molecules! Molecules are secreted through the cell membrane into the extracellular environment. VAI eventually diffuses back into cells once a threshold concentration (~10 μg/ml) has been reached. VAI is a transcription factor that increases the RNA polymerase affinity for the lux promoter. Once RNA polymerase has bound the lux genes are transcribed. lux I = VAI synthetase, makes more VAI. lux R = produces Lux R protein which binds to VAI. The luxR-VAI complex can bind to the operator and increase the RNA polymerase affinity again.
The first few VAI molecules help the RNA polymerase bind and start the lux operon, after which the main regulatory protein is the luxR-VAI complex = “autoinduction.” luxR has its own promoter and is not part of the lux operon.
Operon:
- luxI = VAI synthetase
- luxA&B = code for subunits of enzyme luciferase (this is the one that makes light!)
- luxC&D&E = code for luciferin (substrate for luciferase)
- Luciferin is oxidised by luciferase which causes flashes of blue and green light.
The ribosomal and transfer RNA operons of E. coli
Genes encoding tRNA and rRNA are transcribed by RNA polymerase (note that in human cells we have a specific subset of RNA polymerase for this task.) rRNA + tRNA = 98% of cell’s total RNA BUT they are encoded by only 1% of genome! Whilst mRNA is very unstable and only exists for a few minutes at a time in the cell tRNA and rRNA are stable throughout generations of cells -> carried over (very energetically costly to make ribosomes so must stay intact for a long time.) tRNA and rRNAs are made from short lived precursor molecules.
tRNA: mature tRNAs (clover leaf structure) have their 5’ and 3’ flanking regions removed before they are involved in translation. In bacteria all 3 genes associated with tRNA production are clustered together into an operon. The primary transcript of the operon undergoes secondary folding to form a long primary tRNA molecule, which is cut in three places by ribonucleases: RNase P specifically. RNaseP digests the phosphodiester bonds, gives three tRNA molecules which still need their flanking regions removed.
The 5’ phosphate flanking region is removed by RNaseP again, an endonuclease (i.e. cleaves internally, in the middle of the strand.) Precursor tRNA strand is thus removed. RNaseD removes the 3’ OH flanking region and is an exonuclease: cleaves phopshodiester bonds one at a time until it reaches the CCA codon (this is the stop codon, all tRNA molecules have this at the end.)
3 gene operon —> transcribed into the primary transcript —> cleaved at three points by RNaseP —> must remove flanking regions via RNaseP [5’ region] and RNaseD [3’ region, leaves CCA]
rRNA: in bacteria, an “rrn” is a ribosomal RNA operon -> E coli has seven! tRNA genes may also be included in these rrns. Each operon gives an output of a “30S RNA primary transcript”, which is cut enzymatically to give:
- 2 tRNA molecules (use RNase D and RNase P to become mature)
- 16S RNA [small subunit of ribosome]
- 23S RNA [large subunit of ribosome]
- 5S RNA [large subunit of ribosome]
All RNA subunits are made in equimolar amounts as they are all transcribed onto a polycistronic mRNA = rrn efficient way of packaging RNA genes to ensure that they are all in the right location (together) in the right concentration (1:1 ratio.) [S= svedborg units (if centrifuged all have specific sedimentation coefficient.)]
This 30S transcript of rRNA makes a huge stem loop structure with complimentary on both sides. This structure is cleaved at points by RNase III to make a functional, mature rRNA.