yasmeenzeena.github.io
L7: RNA Polymerase
Aims of this lecture:
- The role of RNA polymerase and the transcription of an RNA molecule from a DNA template. Termination of transcription in the bacterium E.coli. Both rho independent and rho dependent termination of transcription will be described.
TRANSCRIPTION
Most transcriptional information we have is based off of the E. coli model. Transcription in bacteria must be done swiftly. This is achieved by running transcription and translation simultaneously: there is no nuclear membrane so mRNA and protein will form at the same time. In eukaryotes the compartmentalisation of the cell by a nuclear membrane stops translation occurring inside the nucleus.
- no post transcription modification in bacteria means no splicing, no poly A tail addition or intron removal (no introns at all.) Bacteria have short life so processes like these must be fast. The moment RNA is transcribed protein synthesis needs to begin as well.
mRNA = messanger, directs the protein synthesis by carrying the genetic code
tRNA = transfer, carries the amino acid in protein synthesis
rRNA = structural component of the ribosome
Role of RNA polymerase: turns DNA into RNA by catalysing the phosphodiester bond between the 5-phosphate and the 3-OH group on the ribose sugar. Pyrophosphate is a biproduct (PiPi, gamma and beta phosphates are released from the nucleotide when the backbone is formed as only the alpha Pi is kept.) Uracil instead of thymine is used (H rather than a methyl, so uracil is simpler.)
RNA polymerase moves along the DNA from 3’->5’, synthesising the new RNA strand in
the 5’->3’ direction.
 RNA synthesis was proved using Sanger sequencing: if the 3’ OH is removed then mRNA
synthesis stops, so the phosphodiester bond must be made using the 3’ OH.
RNA synthesis was proved using Sanger sequencing: if the 3’ OH is removed then mRNA
synthesis stops, so the phosphodiester bond must be made using the 3’ OH.
RNA Polymerase Subunits
- 2 indentical alpha units –> core
- beta unit –> core
- beta prime unit –> core
- these all make up the holoenzyme
- sigma factor
These were first visualised by cellulose acetate chromatography, a gel filtration precursor that also separates by size.
RNA polymerase MUST: 1. recognise the binding site (as RNA polymerase binds nucleotides together without a primer this is key!) 2. catalyse the phosphodiester formation between the nucleotides 3. recognise the end of the gene. Binding: sigma subunit recognises and binds to the promoter sequence at the beginning of the gene (upsteam, - value.)
Binding: sigma subunit recognises and binds to the promoter sequence at the beginning
of the gene (upsteam, - value.)
+1 = first base to be transcribed upstream = to the LHS,
negative numbers (promoter and regulation area)
downstream = to the RHS, rest of the gene

sigma 70 means that the sigma unit Mr = 70000
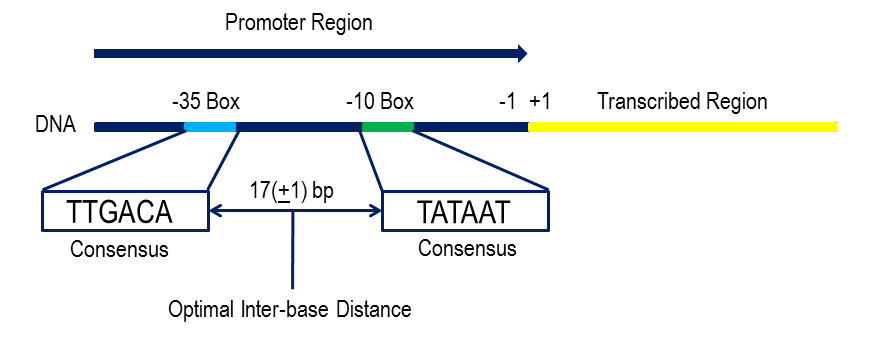
The sigma 70 subunit recognises the -35 and the -10 upstream regions of DNA: these make up the promoter. -35 is the recognition site, with a complimentary DNA sequence to sigma so it can hybridise. -10 region helps the RNA polymerase orient itself and stabilises the a2, b and b’ subunits around the DNA strand. -10 region is AT rich so easily unwinds. Thus, once -35 binding occurs other subunits may also bind.
Most E. coli promoters have a consensus sequence whereby the sigma factor binding
sites are the same! -35 has a TTGACAT sequence, and ~17 bp towards the +1 start site
the -10 region has a sequence of TATAAT. -35 and -10 regions are close by so that the
DNA doesn’t twist around the protein by stays linearised.
A specific experiment was designed to find exactly where the binding site was:
- Find the assumed binding site and make it radioactive, split into two test tubes
- Add RNA poly to the second test tube
- Add DNase 1, an enzyme that digests phosphodiester bonds in DNA
- DNase1 cannot digest the phosphodiester bonds that are protected by RNA poly i.e. no DNA digested where the RNA poly has bound
- Run a gel, the fragment will show where the binding site is Gel was run in a “maxim and gilbert” fashion i.e. chemical cleavage method, lanes are G, G&A, C, C&T (so a mark at G and G&A at the same level suggests a G, and a mark only at G&A is an A.)
rRNA genes are slightly different as they also contain an UP element, which binds another protein to speed up transcription. This makes their promoters stronger.
Weaker promoters can occur when the promoter (-35 and -10 regions) have slightly different sequences to the conserved -> deviations weaken the sigma binding. This can actually aid regulation of a promoter e.g. lac operon, weakness allows a biological switch like lactose to turn it on/off.
B subunit role = nucleotide binding and catalytic activity
B’ role = DNA binding
a role = assembly of the core enzyme (holoenzyme) and aids promoter recognition
RNA polymerase begins situated on the DNA in the closed complex, but once the RNA chain is 8 nucleotides long the sigma factor dissociates and the complex is in the open form. The DNA is unwound, a transcription bubble forms, the 3’->5’ DNA strand acts as the template strand for RNA synthesis.

Detailed mechanism of RNA poly binding: http://proteopedia.org/wiki/index.php/Beta-Prime_Subunit_of_Bacterial_RNA_Polymerase
Termination of transcription: Rho independent [intrinsic termination]
A terminator sequence on the gene must be recognised by the RNA polymerase. This region is characterised with inverted repeats which once transcribed create a hairpin loop. After the hairpin loop there is a U rich section at the end which is the proper termination region. Hairpins are usually made of CGs are these are harder to melt.

The hairpin loop is a very strong structure and the mRNA is left with a UUU section directly after, which is hybridised to the A section of DNA. A=U is much weaker than CG and so the RNA:DNA association is relying on a very weak interaction to hold the complex together. The structure collapses as it is no longer energetically stable!
Termination of transcription: Rho dependent
Rho dependent termination is less common in bacteria. Rho is a hexamer protein (i.e. 6 identical subunits.) Only binds once the mRNA is translated (remember these occur together in bacteria!) and the ribosome has left, has an ATPase activity but mechanism is unknown. Terminators are C-rich and ~40 nucleotides long. The ribosome gets to a translational STOP codon and dissociates, which is when the Rho protein binds to the end of the molecule.
Not all of the downstream after the +1 site is translated from the mRNA strand: the leader and the trailer are not. Therefore, the hairpin structure does not interfere with translation as it is in the trailer section.

- rho is a hexamer protein: 6 identical monomer subunits
- binds to RNA after rho translated (stop codon reached) and ribosome left
- rho binds at recognition site on RNA
- has ATPase activity: used to translocate along RNA
- acts like helicase: breaks RNA away from DNA
- terminators are C-rich and ~40 nucleotides long
- mechanism of termination not clear