yasmeenzeena.github.io
L13: Using DNA to make proteins
Expression systems: bacteria, insect cells, mammalian cells
Protein is scarce —> express protein through cDNA (convert mRNA to cDNA, thus only exonic DNA used.) This allows you to mutate the cDNA.
cDNA must have:
- ATG start (translation initiation) this is the DNA version of AUG remember
- TAA/TAG/TGA stop codon
- inducible promotor
when expressing proteins we need inducible promotors to control when genes are switched on/off. This is because large amounts of protein in the cell can become toxic -> inclusion bodies are formed as the protein precipitates out.
An expression vector for bacteria would thus have:
- inducible promotor upstream of the recombinant gene
- a ribosome binding site specific to prokaryotes
- a MCS with: a recombinant gene made from the mRNA
- a transcription terminator e.g. rho independent
Inducible promoter
- Must have strong binding to the RNA polymerase. The hybrid trc is an example: features the -10 box from the Lac promotor, so binds the LacI repressor (hence inducible) and the -35 box of the Trp promoter, which binds strongly. This trc hybrid is inducible by adding IPTG, which binds preferentially to the LacI repressor and unbinds it from the promoter. IPTG = inducer molecule.
- add a plasmid with the cDNA and a t7 phage promoter insert. The bacteriophage promoter = more control! Add another plasmid that codes for the t7 RNA polymerase -> if we stitch this plasmid on/off we can control transcription of the cDNA via the t7 phage promoter. The second plasmid, with the t7 RNA polymerase, is inducible via the lacI repressor and IPTG.
Issues of using bacteria:
- no post translational modifications, so glycosylation and other effect are lost (e.g. phosphorylation, other Golgi stuff.)
- often inclusion bodies form as the proteins are insoluble / too many are made
- incorrect folding due to prokaryotic environment
- little activity (folding consequence)
Use insect cells as these are eukaryotic! Use a virus (baculovirus) to infect the insect cells with your cDNA of choice, result = correct folding, correct activity and post translational processing (goes to the correct organelle as ER signalling onwards works) & a high yield. Insect hosts are most commonly:
Fall army worm – Spodoptera frugiperda Sf9 Or Cabbage looper – Trichoplusia ni Tni
The baculovirus used to infect cells is about 130kb but difficult to manipulate in a test tube -> few unique restriction sites, hard to get cDNA in (very large insertion.) So instead of using restriction/ligation enzymes use transposition within the bacterial cell:
- get a small donor plasmid and insert the recombined gene of interest
- this plasmid will need two transposition sites straddling the gene: Tn7R
- the plasmid must also have a promoter recognisable by insect cell ran polymerase (eventually this DNA ends up in the insect cell)
- e.g. polyhedrin promoter
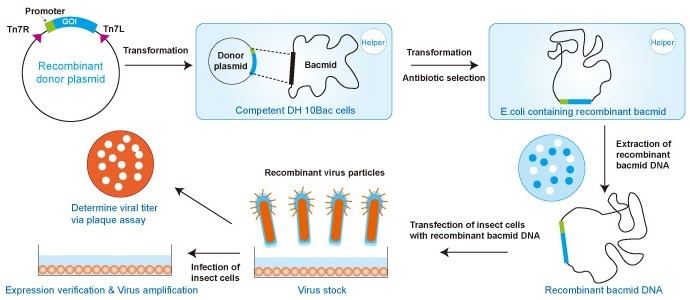
transposition occurs in a bacterial cell -> get competent e coli cells that have the baculovirus genome within! The lacZ gene has been added with a transposition site into the baculovirus genome. LacZ allows blue white colour selection = bacmin molecule
A helper vector is also in the e coli, containing proteins needed for transposition.
Once the viral particles are achieved you can isolate them and infect the insect cells with the virus! Insect cells will lyse after 72 hours so experiment is short term only.
Issues of using mammalian cells:
- expensive
- poor yield
- BUT:
- correct folding
- correct post-translational processes
- correct targeting
- correct activity
Shuttle vectors
Exist in both eukaryotes and prokaryotes, thus have two origins of replication to allow replication in both systems. REMEMBER the polyA signal must be added into the recombinant gene as the cDNA will not have this signal natively. Two different antibiotic selectable markers are used - one for bacteria, one of eukaryotes e.g. ampr and neor.
Transient transfection = plasmid will eventually be degraded after 72h as it is not stable. Transfection achieved through lipid treatment/electroshock to get the plasmid into the cell. Lipid treatment = coat the DNA of the plasmid in lipids so that hopefully straight diffusion can occur (lipofection.) Plasmids must travel to the nucleus to get the RNA polymerase to transcribe the cDNA.
Stable transfection = plasmid is able to integrate into the host genome, thus will not be degraded. The protein in stably expressed in the cell.
Uses: define amino acid sequence for organelle import/protein trafficking -> add a suspected signal sequence to a protein and see where it goes.
e.g. mitochondrial protein import sequence defined by chimaeric proteins:
add a signal sequence for the matrix to a cytosolic sequence via a linker (hence chimaeric protein made) -> where does it go? The spacer (51aa) is added to allow the protein to span the mitochondrial membrane (unknown what the protein will do.)
It was found that the protein entered the matrix. The signal sequence and the spacer sequence were cleaved once in the matrix, leaving the DHFR in the matrix folded natively. The protein can only enter the matrix is unfolded of course. The drug methotrexate can be used as it folds the protein irreversibly. If methotrexate is bound to DHFR, the DHFR cannot enter the matrix -> the signal sequence will enter the matrix and be cleaved but the bulk of the DHFR and the spacer will remain on the cytosolic face. Gets stuck in the TOM and TIM channels.
Use GFP to track mutant protein location.
Oligonucleotide site-directed mutagenesis
Make a mutagenic primer, anneal it to your single stranded DNA template, let DNA polymerase extend it. Use Pfu, a DNA ploymerase proofreader, to ensure that there are no other mutations. Denature the dsDNA and reanneal it -> four possible combinations will occur (parent2, mutation2, parent-mutation, mutation-parent.) Mutant strands lack methylation, so if we add Dpn1 to digest the methylated strands the only viable dsDNA left will be mutagenic on both strands.
Thus we need:
- Nucleotide with the mutation
- ssDNA template
- DNA polymerase
- normal nucleotides